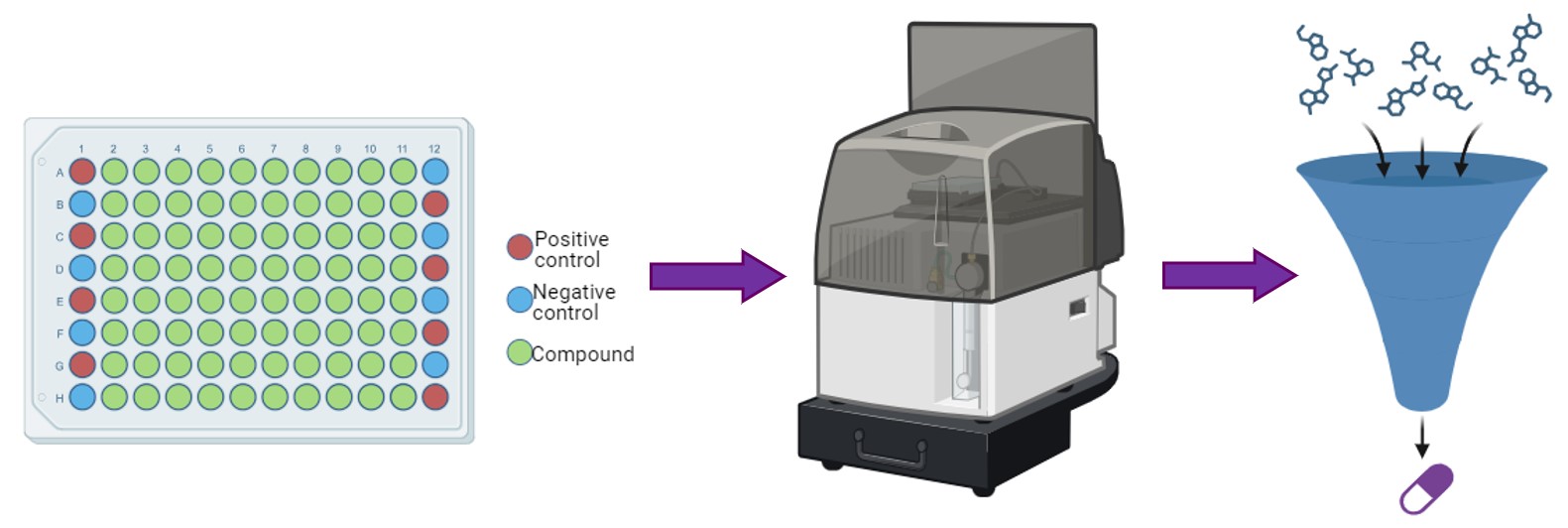
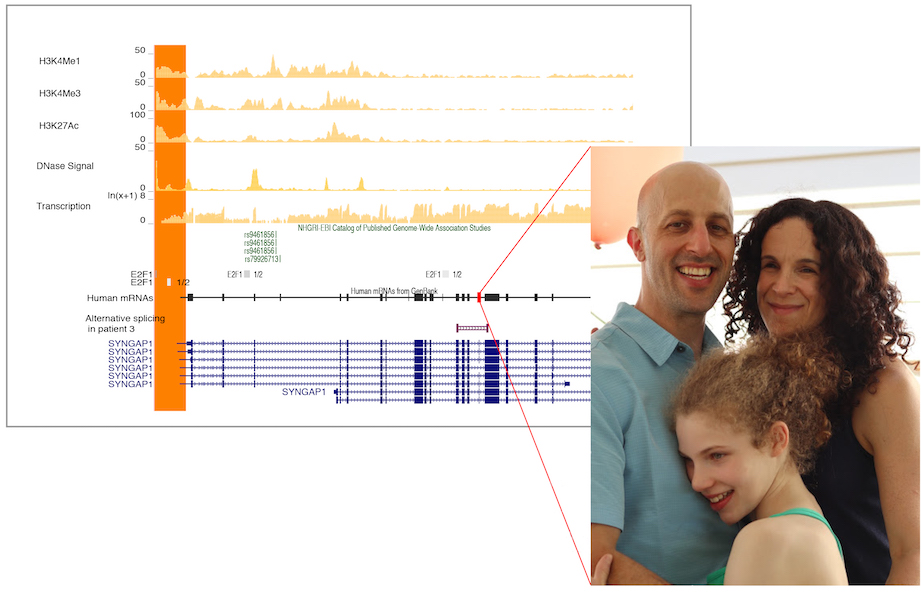
The ability to generate human induced pluripotent stem cells (hiPSCs), which are biochemically and morphologically like embryonic stem cells, is a paradigm shift in human disease modeling. Now researchers can generate hiPSCs from non-disease persons as well as from individuals with disease. This is particularly powerful for genetic disorders because hiPSCs – like embryonic stem cells – can differentiate into any cell type in the body. Therefore, we can use neurons derived from patient hiPSCs and compare them to non-disease neurons to identify disease-specific molecular and physiological phenotypes. These phenotypes serve as benchmarks to test the effectiveness of newly developed therapies.
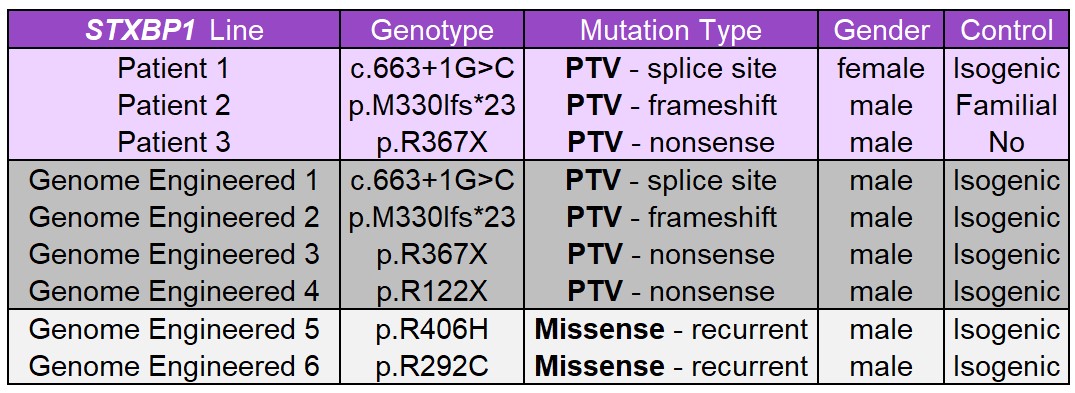
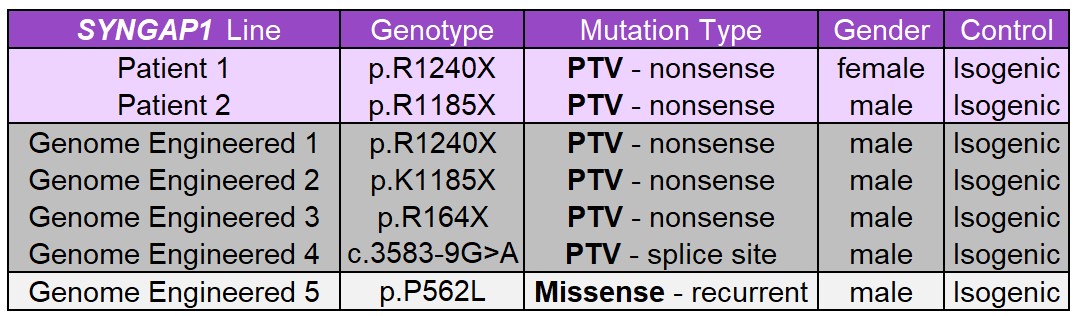
We are developing a panel of STXBP1 and SYNGAP hiPSC lines derived from patients with different pathological variants. We are also using genomic engineering (CRISPR-Cas9) to generate an allelic series of several variants from each gene on a uniform non-disease genetic background, which will enable us to study genotype-phenotype relationships in a more controlled manner. Both types of hiPSC models will permit critically important proof-of-concept studies in human, clinically relevant, cell types.
Dr Deborah French is the lead for Human Stem Cell Model Development at ENDD.
Humanized mouse models of STXBP1 and SYNGAP1 disorders
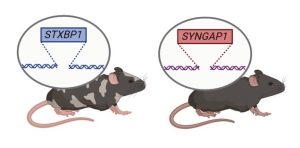
Species differences between rodents and humans have likely played a large role decreased predictive value of rodentmodels for therapeutic development. Many of the therapies we are developing at ENDD require the use of human gene/DNA sequences. Therefore, we have developed humanized mouse of models of STXBP1 and SYNGAP1 where the mouse gene and flanking sequences have been replaced with the human gene and its flanking regulatory sequences. These models will enable the testing of human gene-targeted therapies in an in vivo context. We have confirmed appropriate gene expression from the human alleles relative to the mouse for the STXBP1 and SYNGAP1 humanized heterozygotes and homozygotes. Both fully humanized models have been deposited at the Jackson Labs – prepublication – to make them immediately available to the research community. They are also available upon request (bolandmi@pennmedicine.upenn.edu).
Crossing the humanized mice to heterozygous knock-out mice for each gene will generate humanized mouse models of STXBP1 and SYNGAP1 haploinsufficiency – the primary indication for both disorders – for the assessment of phenotypic rescue after therapeutic intervention. We are also developing humanized mouse lines with specific missense variants (together with corresponding hiPSC lines) for both STXBP1 and SYNGAP1. Once completed, these will also be made available.